Innovation Boosts Study of Fragile Biological Samples at SLAC's X-ray Laser
Researchers at SLAC have found a simple new way to study very delicate biological samples – like proteins at work in photosynthesis and components of protein-making machines called ribosomes – at the atomic scale using SLAC's X-ray laser.
Researchers at the Department of Energy's SLAC National Accelerator Laboratory have found a simple new way to study very delicate biological samples – like proteins at work in photosynthesis and components of protein-making machines called ribosomes – at the atomic scale using SLAC's X-ray laser.
Hasan DeMirci, a SLAC scientist with the Stanford PULSE Institute who teamed up with graduate student Raymond Sierra on the new system, has been using the Linac Coherent Light Source (LCLS) X-ray laser – a DOE Office of Science User Facility – to zero in on the details of ribosomes at work. In addition to their universal role in deciphering the genetic code to build proteins, ribosomes are also important targets for antibiotic treatments.
It is difficult to form ribosomes into crystals so they can be studied with X-rays because they are very fragile. The new system sprang from a desire to better preserve the ribosome crystals.
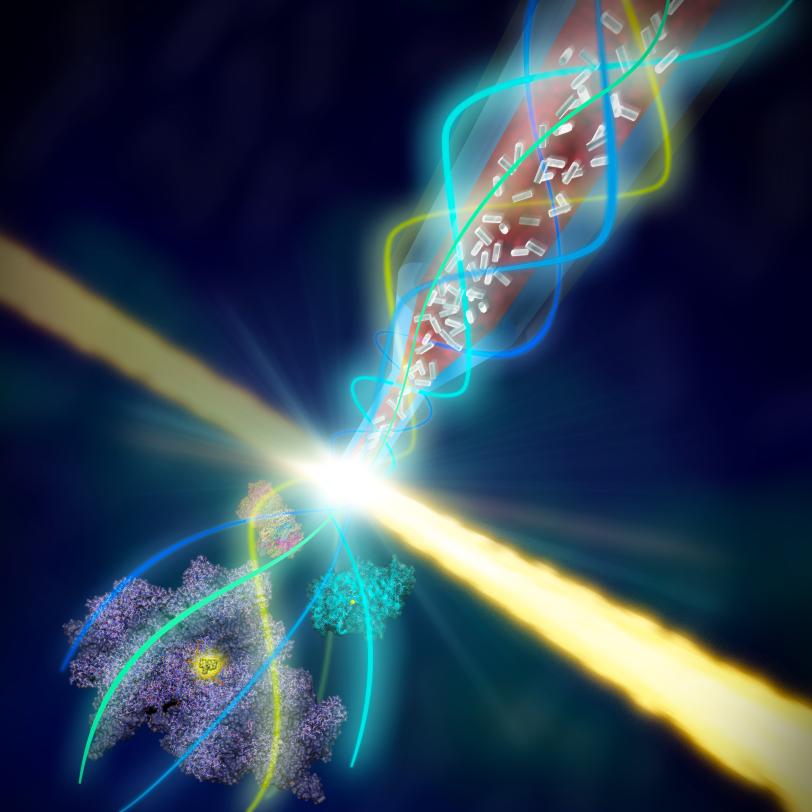
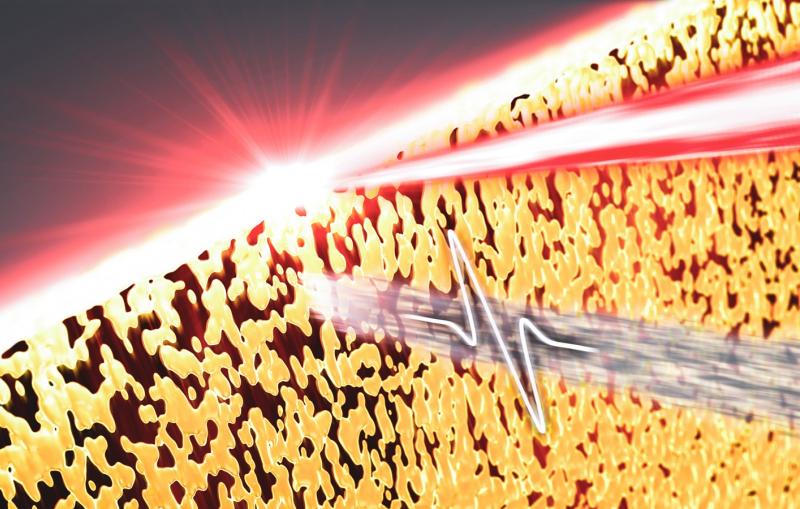
The research team did this by keeping the tiny crystals in the same solution they were grown in at temperatures approaching those in their natural environment, and by finding a more gentle way to deliver or "inject" them into a vacuum chamber, where they are struck by LCLS X-ray pulses.
One Stream Protects Another
The new system, dubbed coMESH, uses low-cost, off-the-shelf components to shape and protect a stream of crystallized proteins with a surrounding stream of electrically charged fluid. It was successfully tested in a 2014 experiment and featured in the Nov. 30 online edition of Nature Methods.
"Our strategy was to come up with an injector that can handle anything, not just ribosomes," DeMirci said. "We are addressing a definite need in the scientific community for a more universal way to deliver samples to LCLS."
In addition to demonstrating that the new system worked, the experiments also gave the scientists a more detailed, 3-D look at how one component of the ribosome binds to an antibiotic called paromomycin that is used to treat parasitic infections. "Now we have a more realistic picture of how this antibiotic interacts with ribosomes at temperatures close to those in their natural environment," DeMirci said.
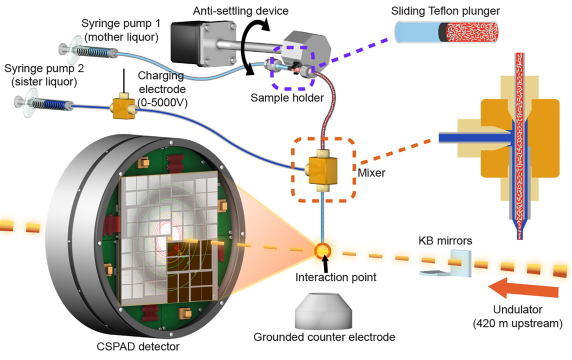
The new system consists of a thin tube, about one-tenth of a millimeter in diameter, inside a slightly larger tube; the sizes can vary based on the dimensions of the crystal samples.
A charging electrode applies low electrical current to the fluid in the larger tube, which focuses the flow to a thin filament. Both tubes end at the same point and the electrical current in the outer fluid greatly narrows both streams of fluid as they emerge from the tubes.
Less Damage and Waste
The system is also designed to waste fewer crystals in experiments than some other sample delivery methods. The thickness and flow rate of the inner stream can be fine-tuned by changing the applied voltage and the width of the tubing to maximize the rate at which X-ray pulses strike the crystals flowing into their path.
DeMirci and Sierra said that based on the 3-D atomic-scale details they were able to see in the ribosome-drug complex and in samples of a photosynthetic protein complex known as photosystem-II, it doesn't appear the voltage damaged the protein structures.
"It's like birds sitting on an electrical wire," DeMirci said.
DeMirci and Sierra said they expect the coMESH system will find wider use by other scientists conducting experiments at LCLS. "We want this to be 'plug-and-play,'" Sierra said, "so all they have to think about during their experiment is collecting data and not troubleshooting sample delivery."
Citation: R.G. Sierra et al. Nature Methods, 30 November 2015 (10.1038/nmeth.3667)
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
SLAC is a multi-program laboratory exploring frontier questions in photon science, astrophysics, particle physics and accelerator research. Located in Menlo Park, Calif., SLAC is operated by Stanford University for the U.S. Department of Energy's Office of Science.
SLAC National Accelerator Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.