Tearing apart a million-dollar microscope – for science
Peter Dahlberg has combined two complex imaging techniques into one. The 2021 Panofsky Fellow adds cryo-ET and biosensors to fluorescence microscopy to give context to high-resolution images of individual proteins in cells.
By Elise Overgaard
Tinkerer though he is, Peter Dahlberg did not spend the last few years tearing apart a $1.5 million microscope just for fun.
Instead, Dahlberg, a staff scientist and 2021 Panofsky Fellow at the DOE's SLAC National Accelerator Laboratory, and his colleagues are aiming to improve some of the world's most powerful microscopes to visualize in minute detail what's happening inside cells.
As a postdoctoral fellow at Stanford, Dahlberg worked with both W.E. Moerner, who shared the 2014 Nobel Prize in chemistry for his part in developing super-resolution fluorescence microscopy, and Wah Chiu, a pioneer in cryogenic electron microscopy (cryo-EM) and its cousin cryogenic electron tomography (cryo-ET). Super-resolution fluorescence microscopy is great for tracking single molecules, such as proteins in a cell, but doesn't reveal what else is going on nearby. Cryo-ET, on the other hand, yields very high-resolution pictures of the cell, but can't pinpoint individual molecules or what they're up to – so Dahlberg is combining the two techniques into one.
"The goal is to keep the best of both worlds," Dahlberg said. "You're retaining the molecular specificity of fluorescence microscopy, so you know who's who, and then you can put it in the context of these high-resolution structures from cryo-ET."
Each advance in perfecting the technique puts scientists closer to understanding the molecular machines that drive things like cell division or parasitic host invasion. But combining techniques requires intensive tinkering with very expensive equipment, which Dahlberg said "is fun and nerve wracking at the same time."
Step one: tearing apart that microscope.
Two methods, one microscope
In fluorescence microscopy, researchers tag an individual molecule, such as a protein, with a smaller molecule that glows when researchers shine a light on it. They can then track the exact location of that singular protein of interest inside a cell under an ordinary – if very high resolution – optical microscope. The problem is, you can’t see what else is near the protein or what else is happening in the cell. It lacks context.
On the other hand, cryogenic electron tomography uses electron microscopes to study flash-frozen samples, such as cells. Although the technique can produce extremely high-resolution pictures of cells, it's impossible to pinpoint a single protein or molecule of interest. It lacks specificity.
In his work, Dahlberg combines the two techniques into one, aptly named "super-resolved cryogenic correlative light and electron tomography." He’ll do both techniques separately, then overlay the resulting images to get a clear view of both the target molecule and its surroundings.
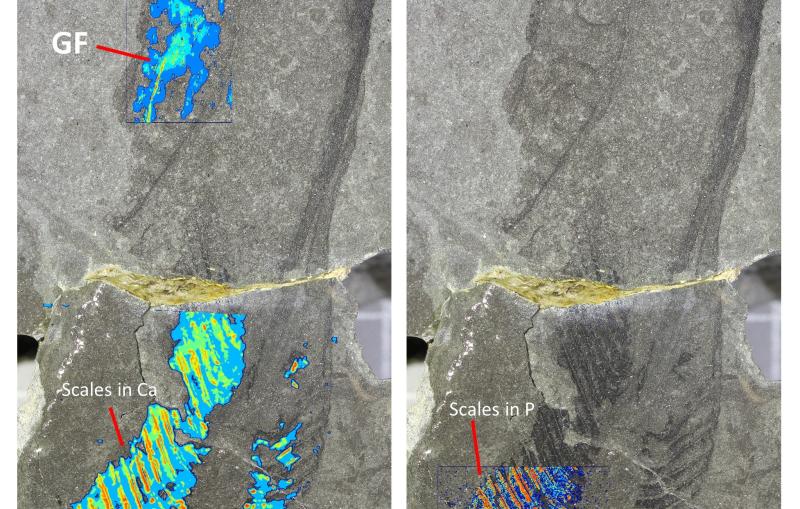
But one problem crops up at the very start of the process. Dahlberg has to drop cells that contain fluorescently-labeled proteins onto a cryo-ET grid – a circular metal mesh only 3 millimeters in diameter – then flash-freeze them so fast that the water on the grid vitrifies, or turns to glass. Once the tiny grid is frozen, it's got to stay frozen – and uncontaminated. The fewer times it's moved, the better.
A second problem is that those frozen cells are thousands of nanometers thick, but the electrons used for cryo-ET can’t penetrate deeper than 200 nanometers.
To solve those problems, Dahlberg has been optimizing a device called a "focused ion beam milling system with an attached scanning electron microscope," or a FIB-SEM. The focused ion beam cuts away cellular material, leaving a very thin slice of frozen cell that cryo-ET electrons can penetrate. Then the scanning electron microscope shoots electrons at the sample and produces those nice, high-resolution cryo-ET images.
But the FIB-SEM doesn't have an optical microscope attached, which means Dahlberg has to move the grid to get fluorescence microscopy data – so Dahlberg gutted his FIB-SEM and added an optical microscope. "Essentially, we just ripped apart this $1.5 million sophisticated instrument to install this integrated light microscope," he said, "and now we have a much, much better system."
The modifications are nearly done, and the team is itching to use it in its full capacity. "I think we're really on the precipice of doing super resolution in the FIB-SEM now," he said. But the team hasn't just been sitting around waiting for the microscope to be finished.
Tinkering with the technique
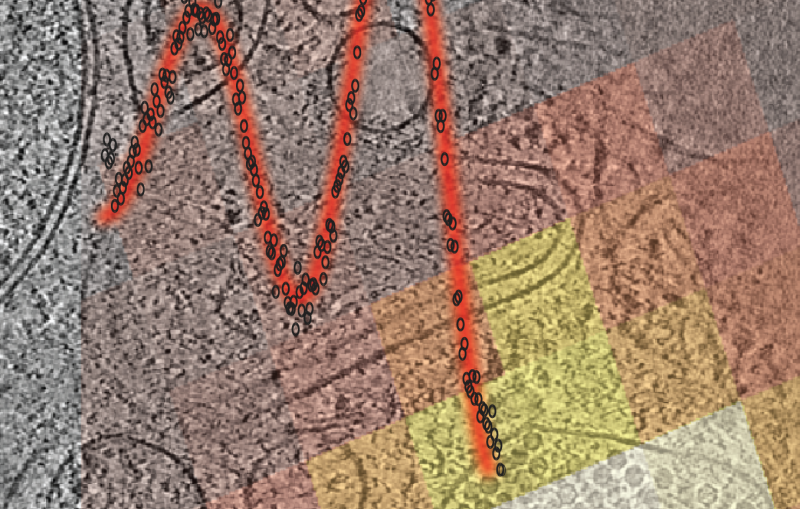
In 2020, Dahlberg used the correlative technique to follow proteins around within Caulobacter crescentus bacterial cells. C. crescentus is interesting because, instead of dividing symmetrically to produce two identical daughter cells, it divides asymmetrically – it produces one mobile cell and one stationary cell. Biologists are interested in that mechanism because it could shed light on similar, if more complex, processes that play out in most human cells.
And Dahlberg thought it was a great system for testing his technique.
Dahlberg and his team tracked two proteins, called SpmX and PopZ, using their technique, which showed exactly where in the cell SpmX and PopZ go during asymmetric cell division. That provided clues to how they interact to perform their individual roles in the bacteria’s life cycle. Perhaps more importantly, the experiment proved that the correlative technique could work.
But it wasn’t perfect. "The fluorescence was really bad," said Dahlberg. "One of the reasons is that we had to keep the excitation light very low. As soon as we shined light on our sample, the samples heated up."
He realized the material the cryo-ET grids were made of was absorbing light and ruining the frozen samples. He wanted to switch to a material with different thermal and optical properties – silver – but nobody makes or sells silver grids.
A few months later, Dahlberg arrived at SLAC. He was surrounded by machine shops and engineers who could make pretty much anything a SLAC scientist dreamt up. So he simply engineered better grids.
But he didn't stop there. Next, he and a team at Stanford led by Davis Perez, a graduate student in W. E. Moerner’s lab, designed a better stage for the light microscope. Now he's engineering different kinds of fluorescent labels – biosensors – to work under cryogenic conditions.
Biosensors are fluorescent molecules that change their emission or excitation properties depending on the local environment, so they'll glow one color in one environment, but a different color in another. "They can be tuned to be sensitive to pH, calcium – you name it," Dahlberg said. "There are hundreds of environmental variables they can be tuned to. So, on top of the specific location and high-resolution structural information, you can also know, was my cell healthy or sick? About to undergo division? At a high ATP concentration? It provides all this extra context."
Dahlberg is determined to keep tinkering with the tools, overcoming technical hurdles one at a time until the technique is optimized and reaches its full potential.
Many hands make light work – and also administrative work
In addition to technical challenges, Dahlberg has tackled the professional challenges of transitioning from post-doctoral to principal investigator life.
"At the end of your post-doc you’re at the pinnacle of your ability to do research, you've got more than a decade of training, you've got great experimental hands, you're ready to take on everything," he said, "but you can't do 20 experiments by yourself at once and have them all work." So he's assembling a rock star team to help.
In 2022, Dahlberg had just one employee – a summer intern. By this fall, he’ll have 10.
The team solves the shortage of hands, but it also pulls Dahlberg away from the bench. And that’s been tough. "So much of the job is no longer about the science directly, he said. "I spend my days thinking about office space, recruiting, writing grants, purchasing problems and getting people funded."
But it's worth it. Dahlberg said it's thrilling to watch his carefully curated team run with ideas. "They do brilliant things you wouldn't have even thought of. Then you walk into the office and hear, 'Oh, I’ve got this cool idea,' and 'I got this exciting result.' You live for those moments."
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
About SLAC
SLAC National Accelerator Laboratory explores how the universe works at the biggest, smallest and fastest scales and invents powerful tools used by researchers around the globe. As world leaders in ultrafast science and bold explorers of the physics of the universe, we forge new ground in understanding our origins and building a healthier and more sustainable future. Our discovery and innovation help develop new materials and chemical processes and open unprecedented views of the cosmos and life’s most delicate machinery. Building on more than 60 years of visionary research, we help shape the future by advancing areas such as quantum technology, scientific computing and the development of next-generation accelerators.
SLAC is operated by Stanford University for the U.S. Department of Energy’s Office of Science. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time.