Researchers reveal how enzyme motions catalyze reactions
What they learned could lead to a better understanding of how antibiotics are broken down in the body, potentially leading to the development of more effective drugs.
In a time-resolved X-ray experiment, researchers uncovered, at atomic resolution and in real time, the previously unknown way that a microbial enzyme breaks down organic compounds.
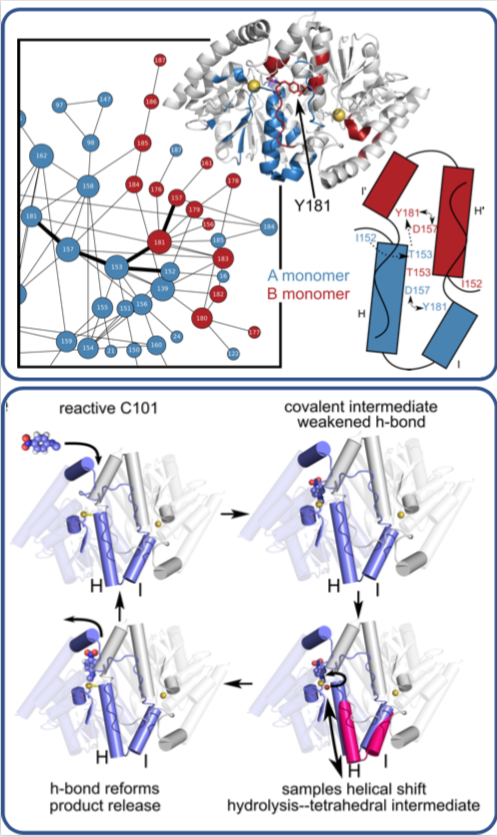
The team, led by Mark Wilson at the University of Nebraska Lincoln (UNL) and Henry van den Bedem at the Department of Energy’s SLAC National Accelerator Laboratory (now at Atomwise Inc.), published their findings last week in the Proceedings of the National Academy of Sciences. What they learned about this enzyme, whose structure is similar to one that is implicated in neurodegenerative diseases such as Parkinson’s, could lead to a better understanding of how antibiotics are broken down by microbes and to the development of more effective drugs.
Previously, the researchers used SLAC’s Stanford Synchrotron Radiation Lightsource (SSRL) to obtain the structure of the enzyme at very low temperatures using X-ray crystallography. In this study, Medhanjali Dasgupta, a UNL graduate student who was the study’s first author, used the Linac Coherent Light Source (LCLS), SLAC’s X-ray laser, to watch the enzyme and its substrate within the crystal move and change as it went through a full catalytic cycle at room temperature.
The scientists used special software, designed by van den Bedem, that is highly sensitive to identifying protein movement from X-ray crystallography data to interpret the results, revealing never-before-seen motions that play a key role in catalyzing complex reactions, such as breaking down antibiotics. Next, the researchers hope to use LCLS to obtain room temperature structures of other enzymes to get a better look at how the motions occurring within them help move along reactions.
SSRL and LCLS are DOE Office of Science user facilities. This work was funded by the DOE Office of Science and the National Institutes of Health, among other sources.
Citation: M. Dasgupta et al., Proceedings of the National Academy of Science, 4 December 2019 (https://doi.org/10.1073/pnas.1901864116)
Contact
For questions or comments, please contact the SLAC Office of Communications at communications@slac.stanford.edu.
SLAC is a vibrant multiprogram laboratory that explores how the universe works at the biggest, smallest and fastest scales and invents powerful tools used by scientists around the globe. With research spanning particle physics, astrophysics and cosmology, materials, chemistry, bio and energy sciences and scientific computing, we help solve real-world problems and advance the interests of the nation.
SLAC is operated by Stanford University for the U.S. Department of Energy’s Office of Science. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.




