Mysterious soil virus gene seen for first time
The protein could play a key role in soil carbon cycling and soil decomposition.
By David Krause
In every handful of soil, there are billions of bacteria, fungi, and viruses, all working to sustain the cycle of life. Understanding how these microorganisms interact with one another helps scientists analyze soil health, soil carbon and nutrient cycling, and even the ways in which dead insects decompose.
Soil viruses contain genes that appear to have some metabolic function, but they are clearly not required for normal viral replication. These genes are called auxiliary metabolic genes (AMGs) and they produce proteins, some of which are enzymes that have a variety of functions. Until now, scientists have wondered whether some AMG proteins play a role in critical soil processes, like carbon cycling. To find out more about soil AMGs, researchers determined the atomic structure of a protein that is expressed by a particular AMG.

Specifically, researchers irradiated fragile crystallized protein samples with high-brightness X-rays generated by the Stanford Synchrotron Radiation Lightsource’s (SSRL) Beam Line 12-2 at the Department of Energy’s (DOE) SLAC National Accelerator Laboratory. The X-rays struck the proteins within the crystal samples, revealing their molecular structures and a bit of the mystery behind their makeup.
AMGs do not, like many viral genes, help a virus replicate. Instead, they encode for a variety of proteins, each with their own predicted function. The AMG that was expressed was a putative enzyme that plays a key role in how soils process and cycle carbon in the biosphere.
“We saw the location of every atom in the viral protein, which helps us figure out how it functions,” Clyde Smith, SSRL senior researcher and co-author, said. “We were amazed to see that the protein resembles known atomic structures of related bacterial and fungal enzyme families, but also contained totally new pieces.”
The detailed atomic structure is unprecedented and reveals for the first time the potential mechanism of this novel enzyme that could play an important role in soil ecology, Janet K. Jansson, chief scientist at the DOE’s Pacific Northwest National Laboratory (PNNL) and co-author, said.
“Our collaboration with SLAC has enabled us to decipher previously unknown functions carried out by soil viruses,” Jansson said.
The research team from SSRL, PNNL, and the Joint Genome Institute (JGI) at the DOE's Lawrence Berkeley National Laboratory, published their results today in Nature Communications.
Breaking down chitin
Researchers think that the viral AMG in the study encodes an enzyme that performs a degradation reaction on chitin. Chitin is the second most abundant carbon biopolymer on the planet after cellulose and is a part of an insect’s exoskeleton and the cell walls of most fungi.
The viral AMG in the study is known as a chitosanase protein, and from sequence analysis was identified as a member of the glycosyl hydrolase GH75 family. This protein could be acting like a garden hoe for the soil – i.e., a tool that helps to prepare soil for vegetables, trees, flowers and all other kinds of life.
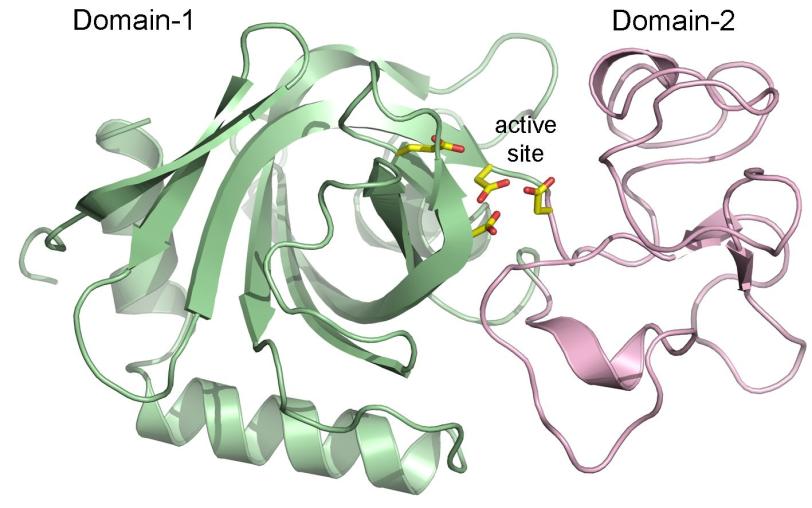
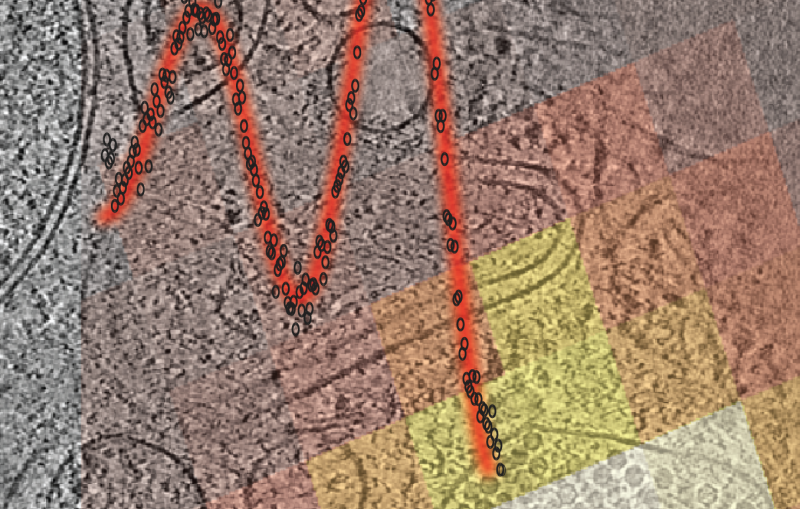
Capturing the atomic structure of the chitosanase protein required more than 5,000 images taken from the crystal samples. Piecing together these images revealed that parts of the protein’s structure resembled a known group of carbohydrate metabolizing enzymes from the glycosyl hydrolase GH45 family. However, the chitosanase protein contained other molecular pieces that did not look like those found in GH45, or in any other known protein structures, which means its role in soil cycling remains open to further studies, Smith said.
“There is a part of the enzyme that is completely new and novel. That’s what’s exciting to me as a structural biologist – to see something we have not seen before, and then try to figure out what its role might be,” Smith said.
Future research could lead to an understanding about why AMGs exist in the first place, since they do not help a virus replicate, Smith said. Additionally, researchers could learn more about other AMGs carried by soil viruses and whether or not they play a functional role in the soil ecosystem.
“One of the big questions coming from this finding is, ‘What in the soil needs that carbon in the chitin?’” Smith said. “Answers to questions like this will lead to a deeper understanding about the interaction of the multitude of microorganisms in the soil, the movement of nutrients and essential molecules, and the overall health of the soil.”
This work was supported by the DOE’s Office of Biological and Environmental Research (BER), JGI, and the DOE’s Environmental Molecular Sciences Laboratory (EMSL). The project was initiated by researchers at PNNL through the soil microbiome SFA funded by BER. It was also supported by a FICUS grant for JGI and EMSL support. The Structural Molecular Biology Program at SSRL is supported by the DOE’s BER and by the National Institutes of Health, and the National Institute of General Medical Sciences.
Citation: Wu, Smith, et al., Nature Communications, YY 19 September 2022 (s41467-022-32993-8)
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
SLAC is a vibrant multiprogram laboratory that explores how the universe works at the biggest, smallest and fastest scales and invents powerful tools used by scientists around the globe. With research spanning particle physics, astrophysics and cosmology, materials, chemistry, bio- and energy sciences and scientific computing, we help solve real-world problems and advance the interests of the nation.
SLAC is operated by Stanford University for the U.S. Department of Energy’s Office of Science. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time.