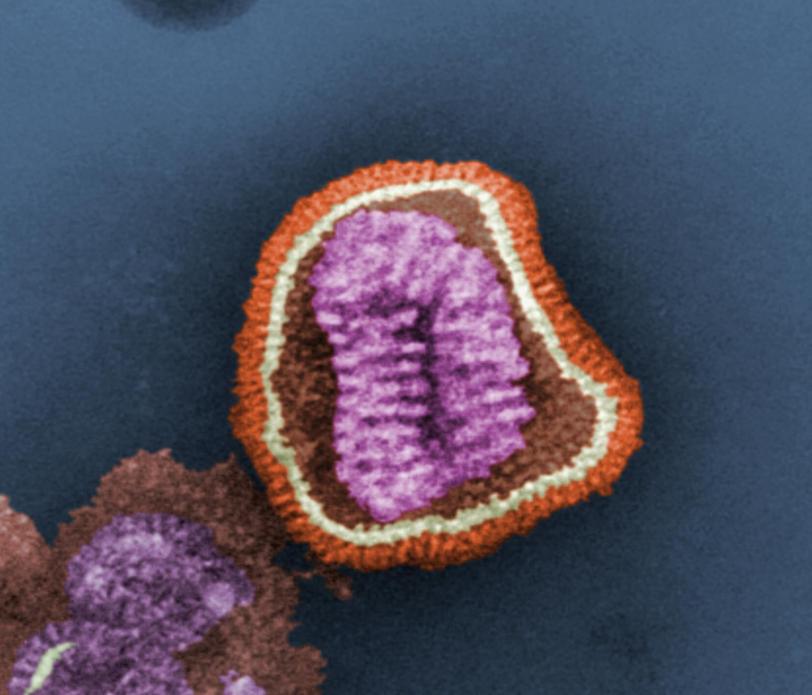
Study May Help Slow the Spread of Flu
X-rays Show How Flu Antibody Binds to Viruses
An important study conducted in part at the Department of Energy’s SLAC National Accelerator Laboratory may lead to new, more effective vaccines and medicines by revealing detailed information about how a flu antibody binds to a wide variety of flu viruses.
The flu virus infects millions of people each year. While for most this results in an unproductive and uncomfortable week or two, the flu also contributes to many deaths in the average flu season. And while vaccines are effective in preventing the flu, they require almost yearly reformulation to keep up with the constantly changing virus.
A team of researchers from The Scripps Research Institute, Fujita Health University and Osaka University studied both samples of flu virus components and an anti-flu antibody. The antibody, called F045-092, was already known to neutralize the flu by connecting to the region of the flu virus that binds to host cells, so it can no longer bind to its target and cause infection.
“There are patches of the virus that are more hypervariable than others,” said Peter Lee, a postdoctoral research associate at The Scripps Research Institute and first author of the paper. “But the flu always binds to host cells within the same region, and so that binding site needs to be functionally conserved. That makes it a site of vulnerability.”
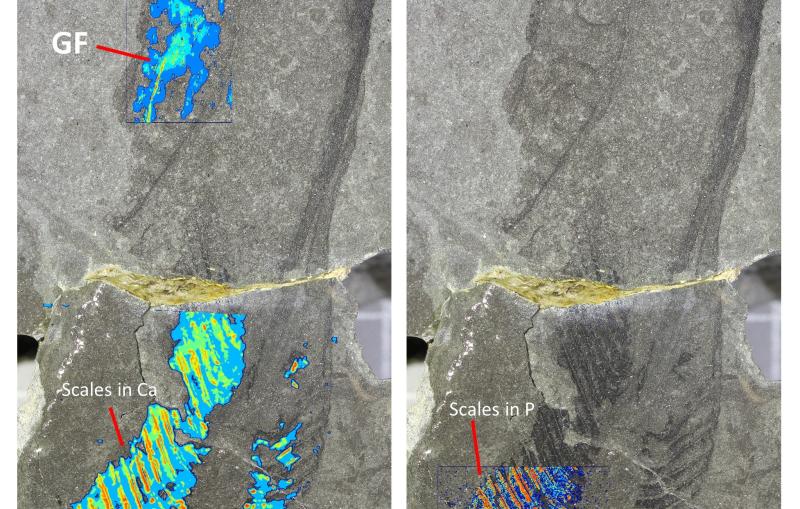
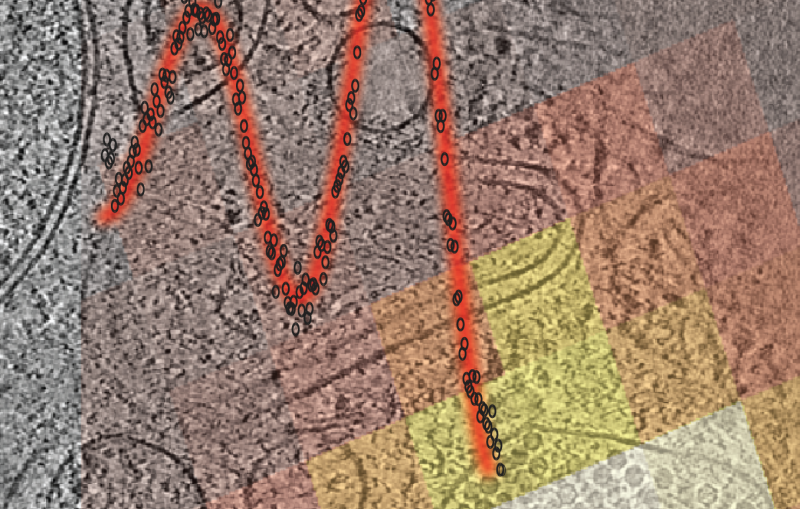
The team used the X-ray beams at SLAC’s Stanford Synchrotron Radiation Lightsource (SSRL) and Argonne National Laboratory’s Advanced Photon Source (APS), both DOE Office of Science User Facilities, to view the structure of the antibody bound to one subtype of the flu virus called H3N2. They discovered that the antibody inserts a loop into the binding site of the virus, which would otherwise attach to a receptor in a host cell. Additional experimental data showed that F045-092 binds a wide variety of strains and subtypes, including all H3 avian and human viruses from 1963 to 2011 that were tested.
This understanding of the antibody's structural details and binding modes offers new insight for future structure-based drug discovery and novel avenues for designing future vaccines.
But the only way to achieve those goals is for many groups of scientists to work together, Lee said. “Our lab is very focused on the structure of the virus and antibodies, while there are lots of other labs focused on everything from small protein design to vaccine design,” he said. “Hopefully we can use this structural information and join together as one big team to tackle the flu.”
SSRL's Structural Molecular Biology program is supported by the National Institutes of Health and the Office of Biological and Environmental Research of the U.S. Department of Energy.
Citation
P. S. Lee, N. Ohshima, et al., Nature Communications 5, 3614 (2014), (10.1038/ncomms4614).
Contact
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
SLAC is a multi-program laboratory exploring frontier questions in photon science, astrophysics, particle physics and accelerator research. Located in Menlo Park, Calif., SLAC is operated by Stanford University for the U.S. Department of Energy's Office of Science.
SLAC National Accelerator Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.
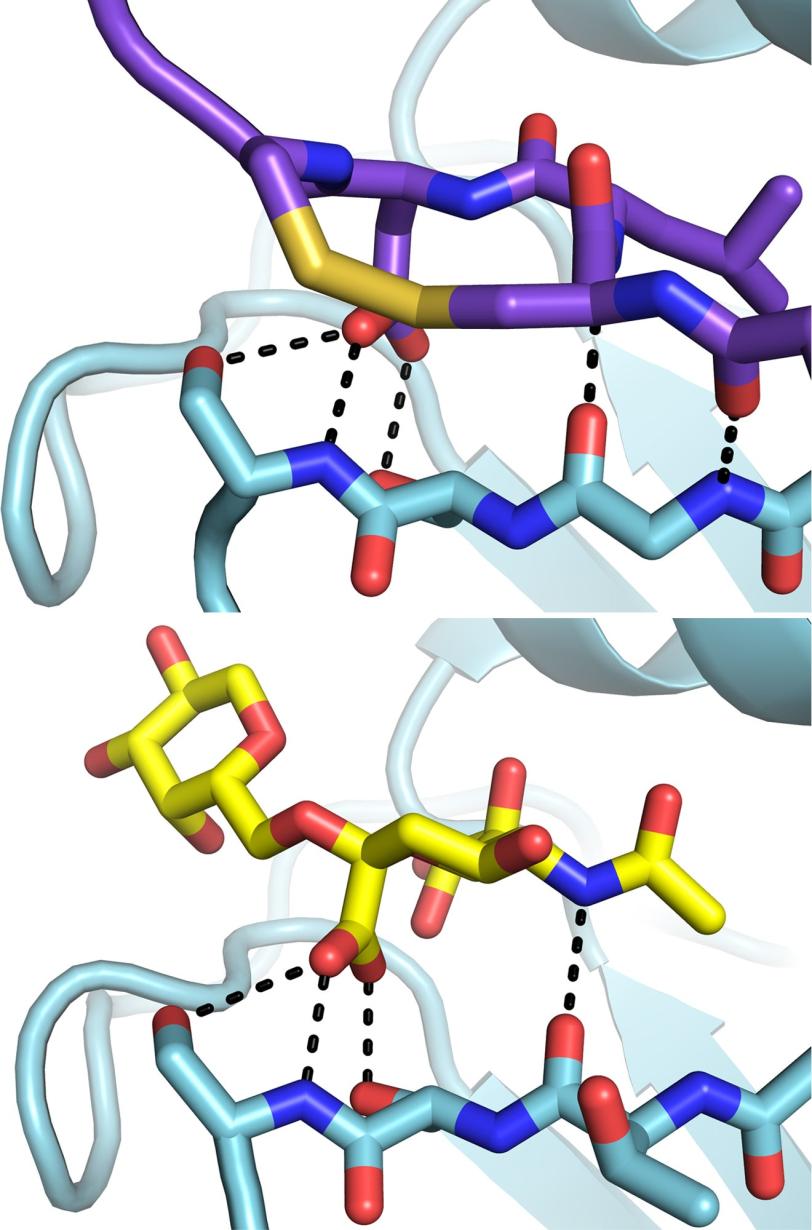
Bottom: Without the antibody present, the flu virus (blue) binds to a host cell receptor (yellow). (Peter Lee et al.)