X-ray Laser Explores New Uses for DNA Building Blocks
The founding father of DNA nanotechnology – a field that forges tiny geometric building blocks from DNA strands – recently came to SLAC to get a new view of these creations using powerful X-ray laser pulses.
By Glenn Roberts Jr.
The founding father of DNA nanotechnology – a field that forges tiny geometric building blocks from DNA strands – recently came to SLAC to get a new view of these creations using powerful X-ray laser pulses.
For decades, Nadrian C. "Ned" Seeman, a chemistry professor at New York University, has studied ways to assemble DNA strands into geometric shapes and 3-D crystals with applications in biology, biocomputing and nanorobotics.
He said the experiment conducted Feb. 7-11 at SLAC's Linac Coherent Light Source enabled his team for the first time to study the DNA structures using smaller crystals in solution at room temperature.
They want to find out whether they can analyze the structure of their samples more precisely in this natural state, as their previous work relied on larger, frozen samples and the freezing process can damage the DNA structures.
"I think we'll get some pretty exciting results," Seeman said during the last shift of the team's LCLS experiment. "I'm very excited by everything I have seen so far."
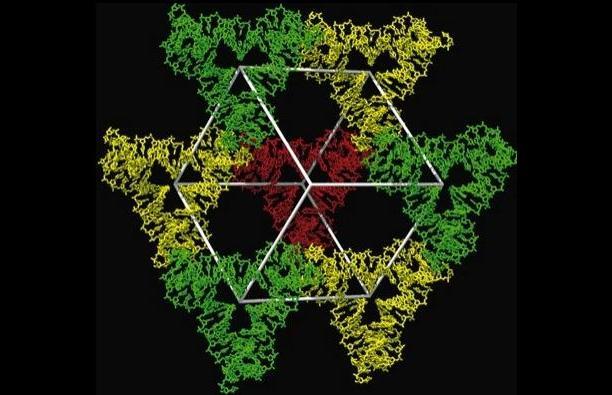
The DNA crystals were suspended in fluid and streamed across the path of the ultrabright, ultrashort LCLS X-ray laser pulses. Detectors captured images, known as diffraction patterns, produced as the X-ray light struck the crystals. The technique is known as X-ray nanocrystallography.
SLAC’s Sebastien Boutet, an instrument scientist at the LCLS Coherent X-ray Imaging Department, said the DNA crystals used in the experiment measured up to about 2-5 microns, or 2-5 thousandths of a millimeter, in size. The crystals were largely triangular and were self-assembled from 3-D DNA objects, forming an ordered lattice. The first-of-its-kind experiment at LCLS involved "lots of trial and error to find the ideal way to prepare the samples," Boutet said.
The engineered structures exploit the natural chemical pairing of DNA to bond small strands of DNA together. The resulting structures can be used to build tiny mechanical boxes and programmable robots for targeting disease, for example.
Researchers can also use DNA engineering as a platform for studying other molecules, such as proteins, that are important to disease research and drug development but are difficult to crystallize, which makes them hard to visualize.
When these proteins are attached to a DNA scaffolding, like salt coating a pretzel, the patterns they form can help scientists analyze their structure.
"The goal, ultimately, is to be able to use this lattice as a crystallization vehicle for things that may not so readily crystallize," Seeman said, "and also for controlling matter, in general, on the nanometer scale."
The ability to form a lattice out of DNA strands, coupled with the fundamental role DNA plays as a biological data storage medium, has also spawned research in DNA-based computing, in which DNA's chemistry and structure are manipulated to store data and carry out computing tasks, performing the functions of magnetic hard drives and silicon chips.
"The key point of DNA is it's got information – it's programmable," Seeman said.
Seeman's research team has previously used synchrotron facilities at Argonne and Brookhaven national laboratories for experiments. He learned about the capabilities at LCLS from Hao Yan, a former student, now at Arizona State University, who has participated in nanocrystallography experiments at LCLS.
Collaborators from ASU, SLAC and research centers in Hamburg and Heidelberg, Germany, participated in the DNA crystallography experiment. Seeman’s team has already submitted a proposal for a follow-up experiment at LCLS.
Contact
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.