Enzyme Created in Test Tube Displays New Structure, Function
Five years ago, a pair of researchers used a clever update on a technique called in vitro evolution – evolution in a test tube – to turn an ordinary protein into an artificial enzyme, a biological catalyst capable of joining two segments of RNA. It was the first time this had ever been done.
By Lori Ann White
Five years ago, a pair of researchers used a clever update on a technique called in vitro evolution – evolution in a test tube – to turn an ordinary protein into an artificial enzyme, a biological catalyst capable of joining two segments of RNA. It was the first time this had ever been done.
Now, a study partially conducted at SLAC’s Stanford Synchrotron Radiation Lightsource has revealed the true extent of their success: not only was the artificial enzyme the first one known to carry out this particular reaction, but it had taken on a new, essentially “primordial” structure never seen in proteins before.
On a practical level, said Seelig, this technique of in vitro directed evolution has the potential to produce new enzymes for a range of applications, such as drug development or biofuel production, without being limited to the familiar ways proteins fold themselves into compact structures.
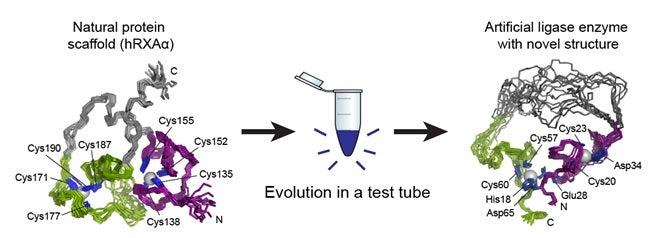
In the original 2007 paper, published in Nature, Jack W. Szostak of the Howard Hughes Medical Institute and Massachusetts General Hospital and Burckhard Seelig, now at the University of Minnesota, explained their process. They started with a protein that showed no enzyme activity, made many copies of it that contained random structural mutations, tested the copies to see if any of the mutations had enzyme-like properties, and selected the most promising candidates to copy and mutate again. Several rounds of this in vitro evolution resulted in the artificial enzyme, called a ligase enzyme. Ligase enzymes forge bonds between molecules and many different types exist, but the artificial enzyme the team created does not seem to have a counterpart in nature.
Though they knew what went into their new enzyme and they knew it worked, the two scientists didn’t yet know why or how. During subsequent investigations into the nature of the artificial enzyme, Seelig and his team of researchers brought samples of it to SSRL. There, Staff Scientist Ritimukta Sarangi used a powerful X-ray spectroscopy technique that, along with nuclear magnetic resonance (NMR) imaging, gave them a good look at the enzyme’s unique structure. What they found, as documented recently in Nature Chemical Biology, was a structure with little resemblance to the original protein – in fact, it had little resemblance to existing enzymes either.
Proteins are made up of long chains of amino acids, and the way these chains fold into a compact structure is vital to the protein’s function. Entire families of enzymes with similar folds have been identified and studied, and one of the most important aspects of studying a new protein is determining how it folds. The researchers found that the artificial enzyme kept a zinc “spine” from the original protein, but discarded that protein’s way of folding and developed its own. What’s more, the artificial enzyme was much more flexible than the parent protein, with a simpler structure than usual for an enzyme.
According to Seelig, the simplified environment of in vitro evolution might be the source of such a primitive structure, and since the enzyme has not been subjected to the billions of years of natural evolution that shaped contemporary enzymes, the way it folds can be viewed as an early or “primordial” fold.
The Structural Molecular Biology program at SSRL is supported by the DOE Office of Biological and Environmental Research, and by the NIH National Institute of General Medical Sciences and the National Center for Research Resources.