SLAC/Stanford researchers discover how a nano-chamber in the cell directs protein folding
The results challenge a 70-year-old theory of how proteins fold in our cells and have profound implications for treating diseases linked to protein misfolding.
By Glennda Chui
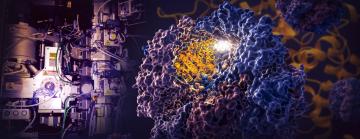
Menlo Park, Calif. — A landmark study by researchers at the Department of Energy’s SLAC National Accelerator Laboratory and Stanford University reveals how a tiny cellular machine called TRiC directs the folding of tubulin, a human protein that is the building block of microtubules that serve as the cell’s scaffolding and transport system.
Until now, scientists thought TRiC and similar machines, known as chaperonins, passively provide an environment conducive to folding, but don’t directly participate in it.
Up to 10% of the proteins in our cells, as well as those in plants and animals, get hands-on help from these little chambers in folding into their final, active shapes, the researchers estimated.
Many of the proteins that fold with the aid of TRiC are intimately linked to human diseases, including certain cancers and neurodegenerative disorders like Parkinson's, Huntington’s and Alzheimer’s diseases, said Stanford Professor Judith Frydman, one of the study’s lead authors.
In fact, she said, a lot of anti-cancer drugs are designed to disrupt tubulin and the microtubules it forms, which are really important for cell division. So targeting the TRiC-assisted tubulin folding process could provide an attractive anti-cancer strategy.
The team reported the results of their decade-long study in a paper published in Cell today.
“This is the most exciting protein structure I have worked on in my 40-year career,” said SLAC/Stanford Professor Wah Chiu, a pioneer in developing and using cryogenic electron microscopy (cryo-EM) and director of SLAC’s cryo-EM and bioimaging division.
“When I met Judith 20 years ago,” he said, “we talked about whether we could see proteins folding. That’s something people have been trying to do for years, and now we have done it.”
The researchers captured four distinct steps in the TRiC-directed folding process at near-atomic resolution with cryo-EM, and confirmed what they saw with biochemical and biophysical analyses.
At the most basic level, Frydman said, this study solves the longstanding enigma of why tubulin can’t fold without TRiC’s assistance: “It really is a game changer in finally bringing a new way to understand how proteins fold in the human cell.”
Folding spaghetti into flowers
Proteins play essential roles in virtually everything a cell does, and finding out how they fold into their final 3D states is one of the most important quests in chemistry and biology.
As Chiu puts it, “A protein starts out as a string of amino acids that looks like spaghetti, but it can’t function until it’s folded into a flower of just the right shape.”
Since the mid-1950s, our picture of how proteins fold has been shaped by experiments done using small proteins by National Institutes of Health researcher Christian Anfinsen. He discovered that if he unfolded a small protein, it would spontaneously spring back into the same shape, and concluded that the directions for doing that were encoded in the protein’s amino acid sequence. Anfinsen shared the 1972 Nobel Prize in chemistry for this discovery.
Thirty years later, researchers discovered that specialized cellular machines help proteins fold. But the prevalent view was that their function was limited to helping proteins carry out their spontaneous folding by protecting them from getting trapped or glomming together.
One type of helper machine, called a chaperonin, contains a barrel-like chamber that hold proteins inside while they fold. TRiC fits into this category.
The TRiC chamber is unique in that it consists of eight different subunits that form two stacked rings. A long, skinny strand of tubulin protein is delivered into the opening of the chamber by a helper molecule shaped like a jellyfish. Then the chamber’s lid closes and folding begins. When it’s done, the lid opens and the finished, folded tubulin leaves.
Since tubulin can’t fold without TRiC, it appeared that TRiC may do more than passively help tubulin spontaneously fold. But how exactly does that work? This new study answers that question and demonstrates that, at least for proteins such as tubulin, the “spontaneous folding” concept does not apply. Instead, TRiC directly orchestrates the folding pathway leading to the correctly shaped protein.
Although recent advances in artificial intelligence, or AI, can predict the finished, folded structure of most proteins, Frydman said, AI doesn’t show how a protein attains its correct shape. This knowledge is fundamental for controlling folding in the cell and developing therapies for folding diseases. To achieve this goal, researchers need to figure out the detailed steps of the folding process as it occurs in the cell.
A cellular chamber takes charge
Ten years ago, Frydman, Chiu and their research teams decided to delve deeper into what goes on in the TRIC chamber.
“Compared to the simpler folding chambers of chaperonins in bacteria, the TRiC in human cells is a very interesting and complicated machine,” Frydman said. “Each of its eight subunits has different properties and presents a distinct surface inside the chamber, and this turns out to be really important.”
The scientists discovered that the inside of this unique chamber directs the folding process in two ways.
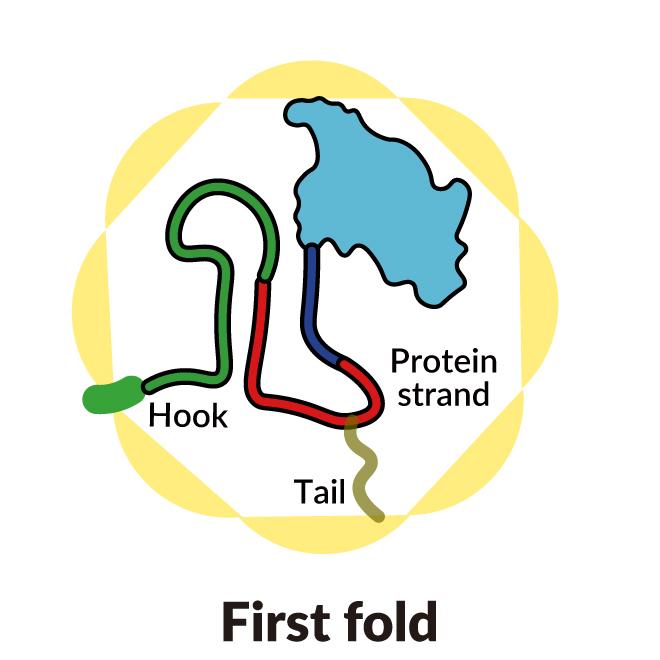
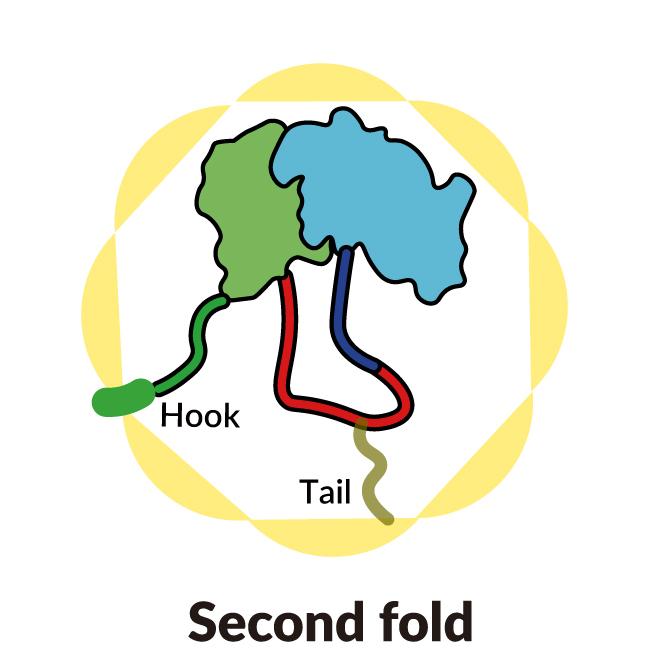
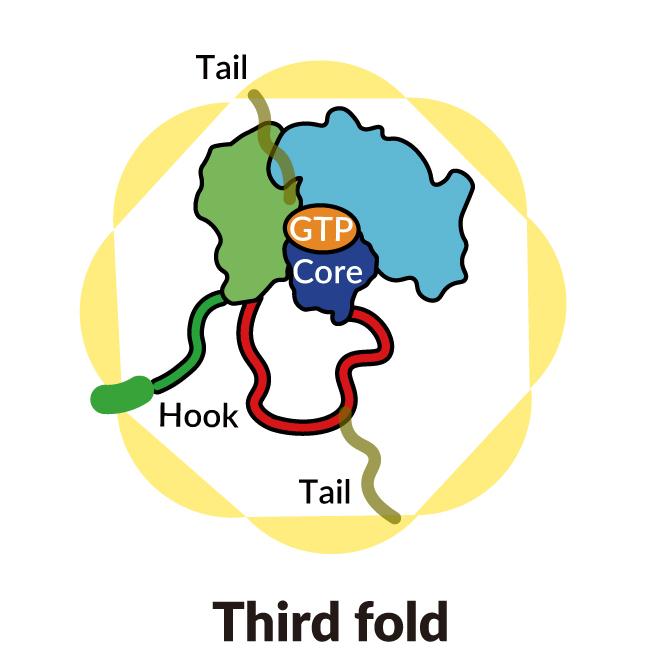
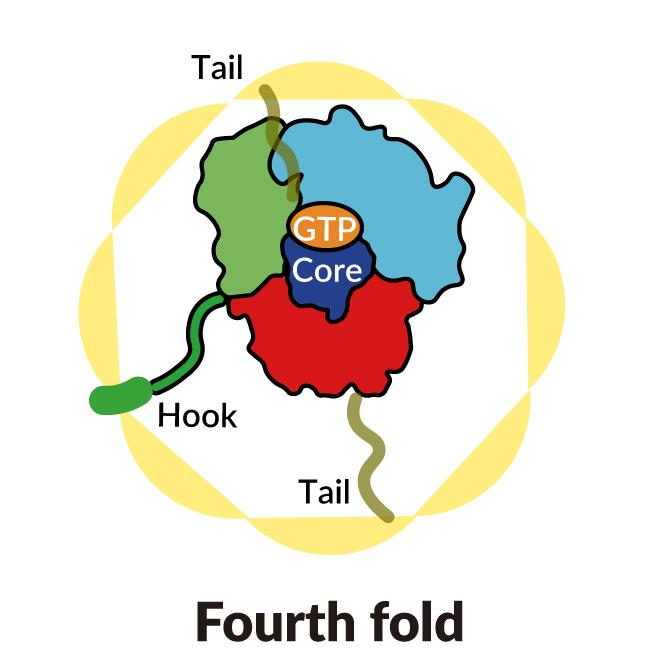
A SLAC-Stanford study revealed four intermediate steps in folding a human protein called tubulin, all directed by the inner walls of a cellular machine called TRiC (yellow). The process starts when a strand of tubulin enters the TRiC chamber. One end (green) hooks into the inner chamber wall; then the other end (light blue) attaches in another spot and folds, followed by the green end and two more folds of the middle sections (dark blue and red). The folding is directed by areas of electrostatic charge on the inner wall and by “tails” of protein dangling from the inner wall, which hold and stabilize the protein in the right configuration for the next step in folding. The protein core (dark blue) contains pockets (orange) where GTP, a molecule that stores and releases energy to power the cell’s work, plugs in. (Yanyan Zhao/Stanford University )
As the chamber’s lid closes over a protein, areas of electrostatic charge appear on its inner walls. They attract oppositely charged parts of the tubulin protein strand and essentially tack them to the wall to create the proper shape and configuration for the next step in folding. Meanwhile, TRiC subunit “tails” that dangle from the chamber wall grab the tubulin protein at specific times and places to anchor and stabilize it.
To start out, one end of the tubulin strand hooks into a little pocket in the wall. Then the other end attaches at a different spot and folds. Now the end that hooked into the wall folds in a way that brings it right next to the first folded area.
In step three, part of the middle section folds to form the core of the protein, along with pockets where GTP, a molecule that stores and releases energy to power the cell’s work, can plug in.
Finally, the remaining protein section folds. The tubulin molecule is now ready for action.
“These structural snapshots of intermediate stages in the folding sequence have never been seen before by cryo-electron microscopy,” Frydman said.
A powerful blend of techniques
Her team confirmed the folding sequence with a challenging series of biochemical and biophysical tests that required years of work.
Interpreting those results allowed the researchers to build a picture of the tubulin’s changing shape as it folds inside the TRiC chamber, which matched the images generated by cryo-EM.
“It’s very powerful to be able to go back and forth between these techniques, because then you can really know that what you see reflects what’s going on in the cell,” Frydman said.
“Science has surprised us with a really interesting solution that I would not have predicted.”
The study also offers clues to understanding how this folding system evolved in eukaryotic cells, which make up plants, animals and humans, but not in simpler cells like those of bacteria and archaea. As proteins became more and more complex to serve the needs of eukaryotic cells, the researchers suggest, at some point they couldn’t fold into the shapes they needed to carry out more complicated jobs without a little assist. Eukaryotic proteins and their chaperonin chamber likely evolved together, possibly starting with the last common ancestor of all the eukaryotic organisms some 2.7 billion years ago.
Due to the complexity of the analyses and the pandemic interlude, the study went on for so long that many of the people who worked on it have moved on to other jobs. They include postdoctoral researchers Daniel Gestaut and Miranda Collier from Frydman’s group, who carried out the biochemical part of the project and pushed it forward, and Yanyan Zhao, Soung-Hun Roh, Boxue Ma, and Greg Pintilie from Chiu’s group, who performed the cryo-EM analyses. Additional contributors included Junsun Park, a student in Roh’s group, and Alexander Leitner from ETH in Zurich, Switzerland.
The work was supported by grants to Wah Chiu and Judith Frydman from the NIH and grants to Soung-Hun Roh, who is now an assistant professor at Seoul National University, from the Korean National Research Foundation and Suh Kyungbae Foundation (SUHF).
Citation: Daniel Gestaut et al., Cell, 8 December 2022 (10.1016/j.cell.2022.11.014)
Press Office Contact: Manuel Gnida, mgnida@slac.stanford.edu, (650) 926-2632
SLAC is a vibrant multiprogram laboratory that explores how the universe works at the biggest, smallest and fastest scales and invents powerful tools used by scientists around the globe. With research spanning particle physics, astrophysics and cosmology, materials, chemistry, bio- and energy sciences and scientific computing, we help solve real-world problems and advance the interests of the nation.
SLAC is operated by Stanford University for the U.S. Department of Energy’s Office of Science. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time.