Protein Structure Solved from Smallest Crystals Yet
Study Reveals High-Res Details of Insect Virus’s Crystal Cocoon
An international team of scientists used an X-ray laser at the Department of Energy’s SLAC National Accelerator Laboratory to determine the structure of an insect virus’s crystalline protein “cocoon.”
The tiny cocoons were by far the smallest protein crystals ever imaged with X-ray crystallography, and the results point the way to using even smaller crystals – or even uncrystallized proteins and other biomolecules – for structure analysis.
Using SLAC’s Linac Coherent Light Source (LCLS), a DOE Office of Science User Facility, the researchers hit nearly 500,000 cocoons with X-ray pulses, creating diffraction patterns in a detector that were compiled to form an image of the cocoon’s structure with a resolution of 0.2 nanometers. The analysis was published in Proceedings of the National Academy of Sciences.
The ability to observe a protein’s structure provides clues about how the molecule behaves, information that can be applied to a wide range of fields such as medicine, agriculture and industrial processes. This study looked at Cydia pomonella granulovirus (CpGV), which infects the caterpillars of the codling moth (Cydia pomonella) and is also used as a biological pesticide. (The virus is harmless to humans.)
“Over the past 50 years, scientists have determined the structures of more than 100,000 proteins,” says Henry Chapman, scientist at Deutsches Elektronen-Synchrotron (DESY) in Germany and leader of the research team. “By far the most important tool for this is X-ray crystallography.”
Normally, scientists performing X-ray crystallography create crystals containing many copies of the protein they want to study. LCLS has allowed them to study much smaller crystals than before, with the ability to capture images with ultrafast X-ray pulses before they are damaged by the intense radiation. This removes a major roadblock to studying proteins that are difficult to make into large crystals.
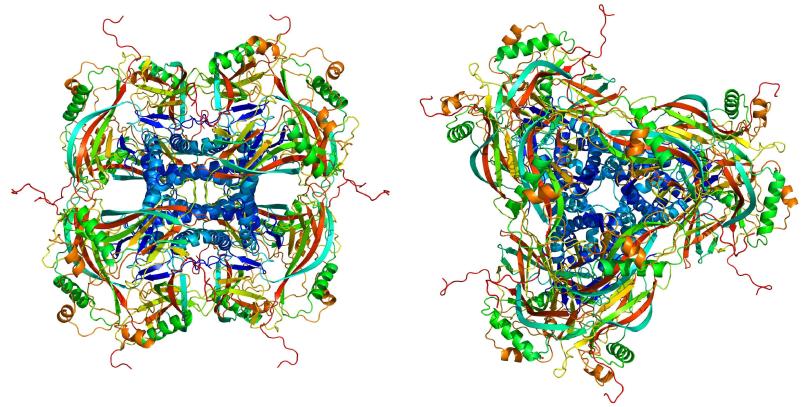
In this case, the virus has a cocoon made of naturally formed crystallized proteins. The crystals each contained about 9,000 copies of the protein, about one-thousand times smaller than crystals used before.
Prior to this result, the typical crystal size used for structural determination at LCLS has been about five microns, with the smallest on the order of one micron [1000 nanometers], says Sebastien Boutet, SLAC scientist and co-author on the paper.
“We’ve pushed the capabilities of LCLS down in the sub-micron region by using extreme focusing that creates a more intense beam on the tiny sample,” Boutet says.
LCLS began the Single Particle Imaging Initiative to work toward atomic-scale imaging for many types of biological samples. To address technical challenges, these efforts have included detailed beam and detector characterization. This effort will benefit from future developments such as building new detectors and upgrading mirrors to improve beam quality.
This paper lays out a path for using free-electron lasers to get structures of proteins and other biomolecules without having to crystallize them at all. In the future, the research team hopes to look at even smaller structures with the same level of detail.
“Simulations based on our measurements suggest that our method can probably be used to determine the structure of even smaller crystals consisting of only hundreds or thousands of molecules,” Chapman says. “This takes us a huge step further towards our goal of analyzing individual molecules.”
Arizona State University, European XFEL, Germany’s Julius Kuhn Institute and Max Planck Institute for Medical Research, University of Auckland in New Zealand, and Switzerland’s University of Basel also participated in the study.
Editor’s note: This article is based on a press release from Deutsches Elektronen-Synchrotron (DESY).
Citation: Gati et al., Proceedings of the National Academy of Sciences, 15 February 2017 (10.1073/pnas.1609243114)
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
SLAC is a multi-program laboratory exploring frontier questions in photon science, astrophysics, particle physics and accelerator research. Located in Menlo Park, Calif., SLAC is operated by Stanford University for the U.S. Department of Energy's Office of Science.
SLAC National Accelerator Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.