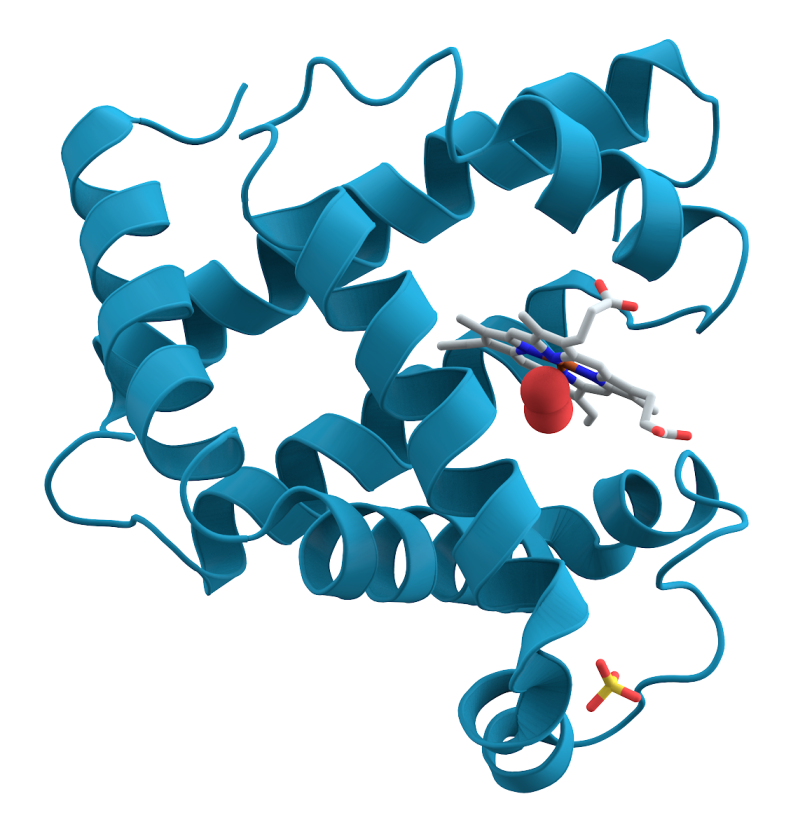Q&A: Biologist Describes Milestone in Watching Proteins Boogie
SLAC's X-ray Laser Enables Direct Images of Ultrafast Structural Changes in Myoglobin
Myoglobin, a protein in muscle tissue that stores oxygen, is what gives meat its red pigment. What is less well-known is that in 1957, it became the first protein molecule whose structure was fully mapped in 3-D. That breakthrough was made possible by a technique called crystallography, in which crystals of a substance are hit with X-rays to produce images that can be used to determine the position of individual atoms. But this early portrait, and the portraits of most all protein structures in the decades that followed, was just a stand-still view. To learn more about how proteins function in the body and how they interact with other molecules, scientists needed a way to watch them shift shape and move.
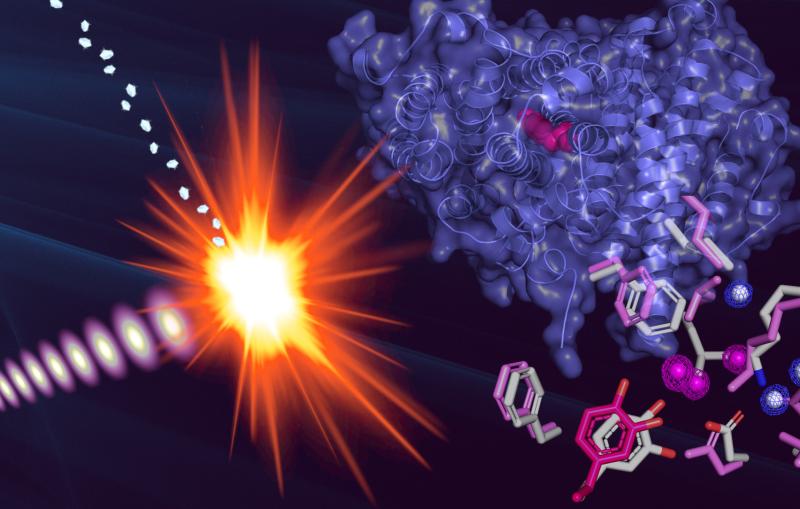
Now, using an X-ray laser at the Department of Energy's SLAC National Accelerator Laboratory, researchers have for the first time directly seen myoglobin move within quadrillionths of a second after a bond breaks and the protein releases a gas molecule. The Linac Coherent Light Source X-ray laser is a DOE Office of Science User Facility, and its short, bright pulses were essential for observing these ultrafast, atomic-scale motions.
Ilme Schlichting of the Max Planck Institute for Medical Research in Germany, a pioneer in structural biology research at LCLS, led the experiment. In this Q&A, she describes how the results, published Sept. 10 in Science Express, mark a milestone in the long-sought goal of watching proteins while they work.
Q: What is unique about this experiment?
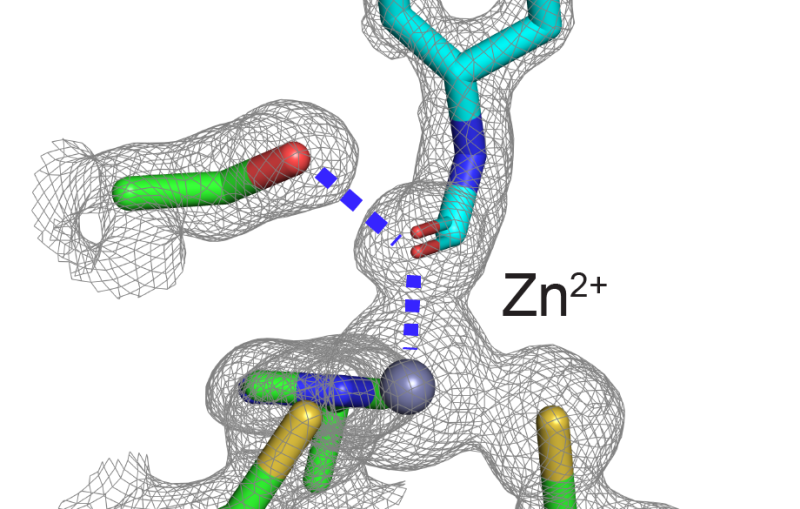
A: It's the very first time we have been able to directly view motions in proteins so soon after a chemical bond breaks. Earlier crystallography studies at synchrotrons, a more conventional type of X-ray facility, were able to probe protein motions as little as 100 picoseconds, or 100 trillionths of a second, after a bond breaks, and other types of X-ray studies had measured this motion in a more indirect way. The question was always, "What happens before that, in the first 100 picoseconds?" Now we actually see the changes happening almost a thousand times faster – down to a few hundred femtoseconds (quadrillionths of a second) after the bond breaks. We see atoms and not just overall movement. It was not possible to do this experiment anywhere except at LCLS or another X-ray free-electron laser, and our results pave the way for studying ultrafast motions in other proteins.
Q: What protein motion did you observe and what it can teach us?
A: We started the reaction by breaking a chemical bond, releasing a carbon monoxide molecule that had been attached to the protein. That breakage induces small local vibrations that quickly cause the whole protein to vibrate. Then, those vibrations drive larger and slower overall protein motions. It’s like shaking a series of connected springs and watching them wobble in different but patterned ways. So what starts as seemingly random vibrations results in parts of the protein moving together in a directed fashion, which is known as a “collective motion.” This shows how a local structural change is transmitted extremely quickly to other parts of a large protein molecule, which has tremendous implications for understanding biochemical reactions in general.
Q: Were you surprised by anything you saw?
A: When we started to do this experiment I really doubted that we would be able to see these vibrations and the collective motions they cause. So for me this was really a pleasant surprise. It presents a new way of looking at molecules, and it’s mind-blowing. We were able to do it because LCLS gives us the ability to collect very accurate data with such precise timing.
These vibrations in the protein had been observed earlier with other techniques, including spectroscopy, but the exact structural motions could only be inferred. Also, no one had been able to directly see these vibrations propagating throughout the entire protein; now we can see that. This and future studies could lead to a more general understanding of how such motions relate to the function of proteins. When you break a single bond, how can that result in a major structural change in the protein? How does it introduce structural changes on the opposite side of the molecule?
Q: What are the next steps for this research?
A: The next step is to look at more complicated systems with maybe a bit more direct biological relevance, such as hemoglobin – myoglobin's bigger cousin that carries oxygen in our blood – and enzymes, which are catalysts that speed up chemical reactions in living things. We want to look at different reactions, different kinds of chemistry, and how these are promoted by protein motions. We want to understand other protein systems from a really basic point of view to see how chemical events promote ultrafast motions and how they result in large structural changes.
In the long term, the development of our ability to see these motions with LCLS will also benefit studies where a reaction might be initiated by something other than a visible light pulse. Because motions are so important to how proteins function, such experiments will not only benefit fundamental biochemistry, but possibly also the design of drugs. In the long run, they could also be important for nanotechnology, by helping in the design of molecular machines based on biological molecules.
Q: What technical advances at LCLS made this latest result possible?
A: The experiment required a constant, reliable stream of samples and sophisticated data processing and analysis methods that were developed specifically for LCLS. Years of improvements and development of liquid jets that delivered the tiny crystals of myoglobin to the X-rays, and improvements in crystallography techniques at LCLS, were put to direct use in this experiment. It was also crucial to have access to a recently developed tool that precisely measured the time between the arrival of the optical laser pulses used to drive the myoglobin reaction and the arrival of the X-ray laser pulses. It is really because of all these developments that experiments such as ours are now possible.
Citation: T.R.M. Barends, et al., Science Express, 10 September 2015 (10.1126/science.aac5492)
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
SLAC is a multi-program laboratory exploring frontier questions in photon science, astrophysics, particle physics and accelerator research. Located in Menlo Park, Calif., SLAC is operated by Stanford University for the U.S. Department of Energy’s Office of Science.
SLAC National Accelerator Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.